pyLiBELa
Weekly Reports
The proposal is in this link.
Week 1
Adapting WRITER class to pyLiBELa
- read the pyPARSER and pyMOL2 classes to understand how it works
- boost WRITER.cpp and WRITER.h
- upload it to github and test in Google Collab
How to generate a shared library with BOOST:
g++ -Wall -Wextra -fPIC -shared / - example.cpp -o example.so -lboost_python37 -lboost_numpy37
Steps I followed to adapt it with boost:
- Know the class funcions and variables by looking at the .h file
- Write the BOOST appendage by the end of the code writing for variables .def_readwrite(“old_variable_name”, & CLASS NAME::”new variable name”), we kept the same name for simplicity for functions .def_write(“old_variable_name”, & CLASS NAME::”new variable name”)
Notes:
- The local machine uses python3.10.6 while Collab uses python3.9.16
- If the function has more than one possible input, adapt it as TNG
- pyLiBELa on github Results:
- The WRITER class is now adapted to pyLiBELa
Tried to Adapt new Mol2 class to pyLiBELa
- see the history log in Source code
- boost new functions and variables
Results:
- The Mol2 class doesn’t work in Google Collab showing the following error message:
In file included from pyMol2.cpp:8:
pyMol2.h:21:10: fatal error: openbabel/obconversion.h: No such file or directory
21 | #include <openbabel/obconversion.h>
| ^~~~~~~~~~~~~~~~~~~~~~~~~~
compilation terminated.
- In the local computer, it still shows the message
In file included from /usr/include/boost/smart_ptr/detail/sp_thread_sleep.hpp:22,
from /usr/include/boost/smart_ptr/detail/yield_k.hpp:23,
from /usr/include/boost/smart_ptr/detail/spinlock_gcc_atomic.hpp:14,
from /usr/include/boost/smart_ptr/detail/spinlock.hpp:42,
from /usr/include/boost/smart_ptr/detail/spinlock_pool.hpp:25,
from /usr/include/boost/smart_ptr/shared_ptr.hpp:29,
from /usr/include/boost/shared_ptr.hpp:17,
from /usr/include/boost/python/converter/shared_ptr_to_python.hpp:12,
from /usr/include/boost/python/converter/arg_to_python.hpp:15,
from /usr/include/boost/python/call.hpp:15,
from /usr/include/boost/python/object_core.hpp:14,
from /usr/include/boost/python/args.hpp:22,
from /usr/include/boost/python.hpp:11,
from pyWRITER.cpp:712:
/usr/include/boost/bind.hpp:36:1: note: ‘#pragma message: The practice of declaring the Bind placeholders (_1, _2, ...) in the global namespace is deprecated. Please use <boost/bind/bind.hpp> + using namespace boost::placeholders, or define BOOST_BIND_GLOBAL_PLACEHOLDERS to retain the current behavior.’
36 | BOOST_PRAGMA_MESSAGE(
| ^~~~~~~~~~~~~~~~~~~~
/usr/include/boost/detail/iterator.hpp:13:1: note: ‘#pragma message: This header is deprecated. Use <iterator> instead.’
13 | BOOST_HEADER_DEPRECATED("<iterator>")
but as the .so archives are compiled, we ignore it.
Week 2
Adapting new Mol2 class to pyLiBELa
- fixing the fatal error adding the g++ flags: -DHAVE_EIGEN; -I/usr/include/openbabel3; -I/usr/include/eigen3
- fixing an error by defining the str variable differently
How to generate the libraries for each class so far:
PARSER
g++ -fPIC -shared -I/usr/include/python3.10 -I/usr/include/openbabel3 -I/usr/include/eigen3 pyPARSER.cpp -o pyPARSER.so -L/usr/lib/x86_64-linux-gnu -lboost_python310
WRITER
g++ -fPIC -shared -DBUILD=0 -I/usr/include/python3.10 -I/usr/include/openbabel3 -I/usr/include/eigen3 pyWRITER.cpp -o pyWRITER.so -L/usr/lib/x86_64-linux-gnu -lboost_python310 -lz
Mol2
g++ -fPIC -shared -DHAVE_EIGEN -I/usr/include/python3.10 -I/usr/include/openbabel3 -I/usr/include/eigen3 pyMol2.cpp -o pyMol2.so -L/usr/lib/x86_64-linux-gnu -lboost_python310 -lz
Adapting Grid class to pyLiBELa
- boost the variables and functions of the class
- adapt the variable info as the variable str of the Mol2 class
Results:
- The Grid class can be turned into a dinamic library in Google Collab and in the local machine through the following command line:
Grid
g++ -fPIC -shared -DBUILD=0 -I/usr/include/python3.10 -I/usr/include/openbabel3 -I/usr/include/eigen3 pyGrid.cpp pyWRITER.cpp -o pyGrid.so -L/usr/lib/x86_64-linux-gnu -lboost_python310 -lz
Week 3
Adapting COORD_MC class to pyLiBELa
- Downloaded the RAND.h from LiBELa’s source code and renamed it to pyRAND.h
- Downloaded the gsl library to local machine
Results:
- The COORD_MC class can be turned into a dinamic library in Google Collab and in the local machine through the following command line:
COORD_MC
g++ -fPIC -shared -I/usr/include/python3.10 -I/usr/include/openbabel3 -I/usr/include/eigen3 pyCOORD_MC.cpp -o pyCOORD_MC.so -L/usr/lib/x86_64-linux-gnu -lboost_python310
Adapting FindHB class to pyLiBELa
Results:
- The FindHB class can be turned into a dinamic library in the local machine
FindHB
g++ -fPIC -shared -I/usr/include/python3.10 -I/usr/include/openbabel3 -I/usr/include/eigen3 pyCOORD_MC.cpp -o pyCOORD_MC.so -L/usr/lib/x86_64-linux-gnu -lboost_python310
- In Google Collab, the code is compiled. But, as the library is imported on python, it shows the following error message appears
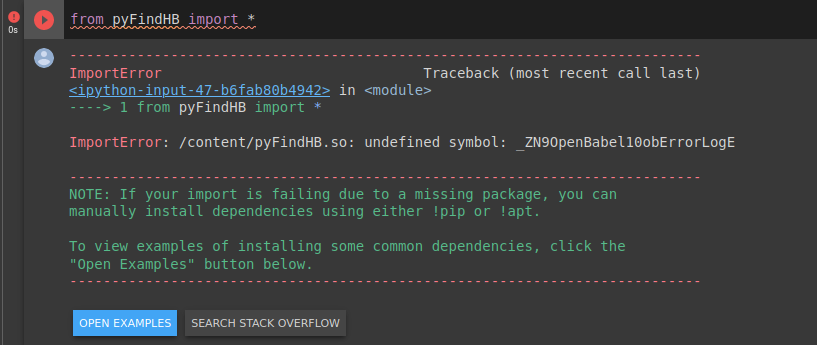
- The Collab error was fixed by adding the -lopenbabel at the end
Creating a makefile
- Learning how to create a makefile
- Creating a makefile to compile all the .so libraries
- Create another one to compile on Google Colab
Results:
- Created a makefile that compiles every dinamic library with by tiping only ‘make’
- The BOOST error appears for every line of code compiled -> fix that in the future
- The makefile to the Google Colab version works, but the libraries pyGrid and pyFindHB can’t be imported.
- The pyGrid library can be imported if you add the following lines to the pyGrid.cpp file:
#include "pyWRITER.h" #include "pyWRITER.cpp"and do the same with the pyFindHB file, but include the pyCOORD_MC.* files instead.
Useful links:
Week 4
Adapting Energy2 class to pyLiBELa
Now that we have a makefile, testing needs to be done in four steps:
- individually in the local machine
g++ -fPIC -shared -I/usr/include/python3.10 -I/usr/include/openbabel3 -I/usr/include/eigen3 pyCLASS.cpp -o pyCLASS.so -L/usr/lib/x86_64-linux-gnu -lboost_python310 - with the other classes through the Makefile
makein the pyLiBELa folder. The files will be transformed in .so dinamnic libraries that will be found in the obj/ directory. To delete everything, you just need to type
make clean - individually in Google Colab
- with the other classes through the Colab Makefile
In this class we had a “subclass” called GridInterpol, written in the .h file as
class Energy2 {
public:
struct GridInterpol{
double vdwA;
double vdwB;
double elec;
double pbsa;
double delphi;
double solv_gauss;
double rec_solv_gauss;
double hb_donor;
double hb_acceptor;
};
so we had to define it in a separate space in the boost appendage
class_<Energy2::GridInterpol>("GridInterpol")
.def_readwrite("vdwA", & Energy2::GridInterpol::vdwA)
.def_readwrite("vdwB", & Energy2::GridInterpol::vdwB)
.def_readwrite("elec", & Energy2::GridInterpol::elec)
.def_readwrite("pbsa", & Energy2::GridInterpol::pbsa)
.def_readwrite("delphi", & Energy2::GridInterpol::delphi)
.def_readwrite("solv_gauss", & Energy2::GridInterpol::solv_gauss)
.def_readwrite("rec_solv_gauss", & Energy2::GridInterpol::rec_solv_gauss)
.def_readwrite("hb_donor", & Energy2::GridInterpol::hb_donor)
.def_readwrite("hb_acceptor", & Energy2::GridInterpol::hb_acceptor)
;
and where it appeared as an argument for trilinear_interpolation(), we put it as Energy2::GridInterpol.
In Colab, if the following error appears
ImportError: /content/pyEnergy2.so: undefined symbol: _ZTI4Grid
just add the corresponding class to the .cpp file. In this case, the Energy2 class depends on the Grid class, so we add to the pyEnergy.cpp file the file
#include "pyGrid.cpp"
and it works.
Results:
- The .so library can be compiled locally
- If we add the new files to the Makefile on the Colab repository, it can be imported in Google Colab.
Notes:
- Shift+Ctrl+V pastes in the terminal
- Ctrl+C interrupts the command in the terminal
Useful links:
- .md files tutorial
Adapting Conformer class to pyLiBELa
Notes:
- The following message appeared
pyConformer.cpp: In member function ‘bool Conformer::generate_conformers_confab(PARSER*, Mol2*, std::string)’: pyConformer.cpp:92:11: error: ‘class OpenBabel::OBForceField’ has no member named ‘DiverseConfGen’ 92 | OBff->DiverseConfGen(0.5, Input->conf_search_trials, 50.0, false); | ^~~~~~~~~~~~~~and the error was fixed by adding to the terminal line a -DHAVE_EIGEN.
Results:
- The Conformer class is ready to be used both in the local machine and in the Google Collab
Updating Makefiles
- Creating an external Makefile for testing in the local machine so that there is only one Makefile, the one from Google Colab.
- Updating the Colab Makefile to be able to compile one function at a time.
Adapting RAND and Gaussian classes to LiBELa
- RAND and Gaussian can be compiled in the local machine and in Google Colab
Listed the dependencies of pyLiBELa library
- The dependencies of each class can be now viewed on the following tables
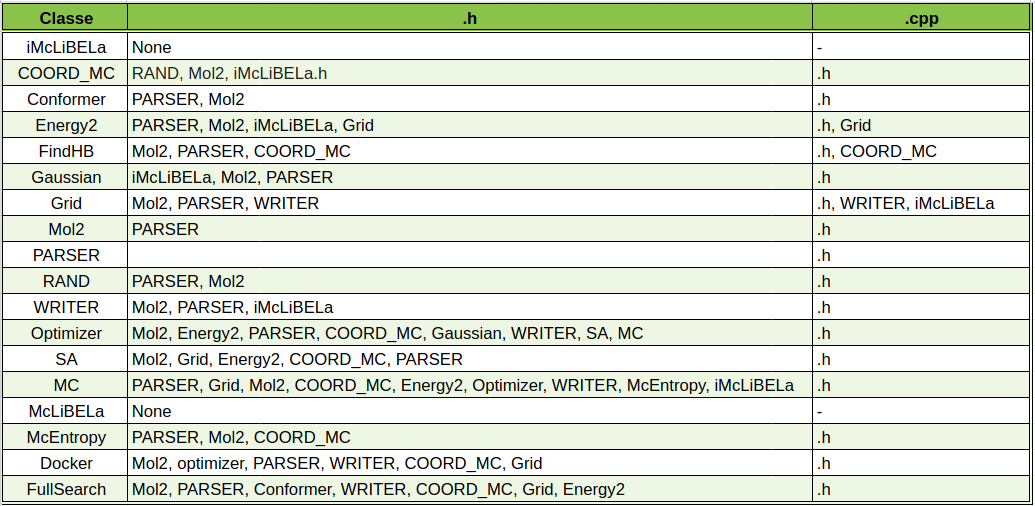
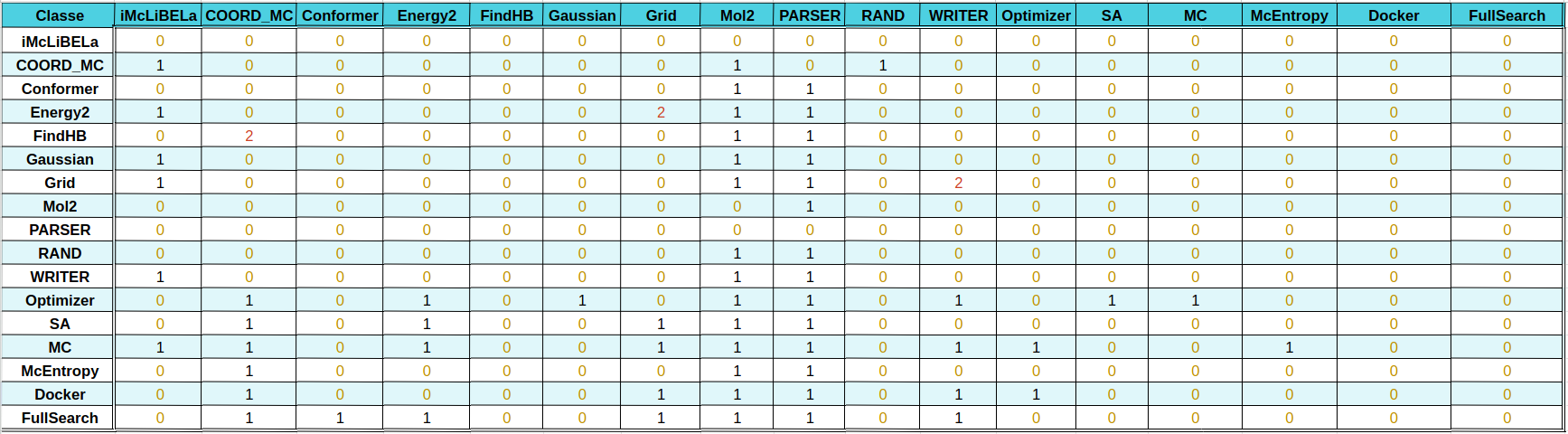 both available at this Google Sheets
both available at this Google Sheets
And they can be turned into a graph
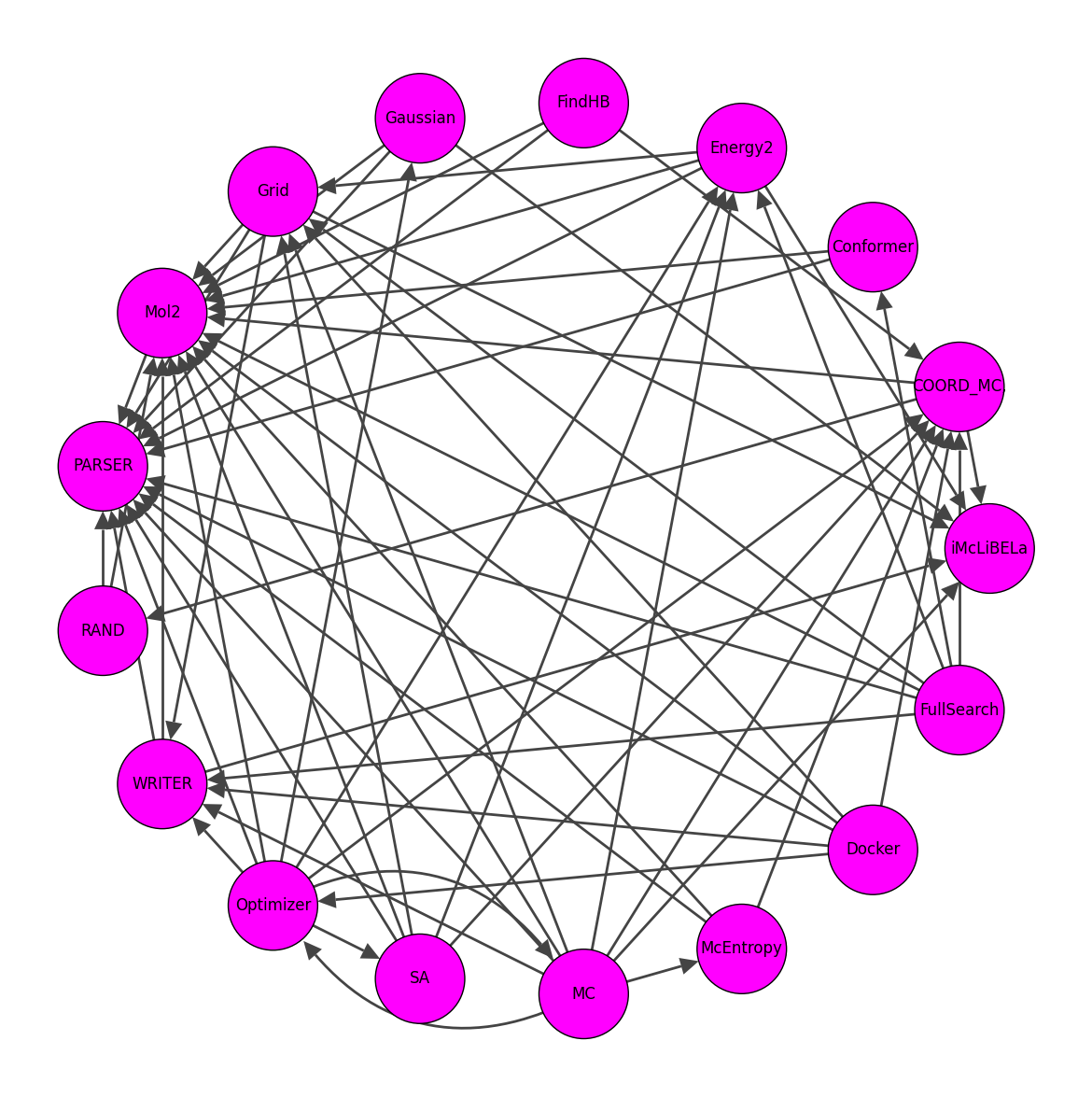 throught this script,
and cleaned to higher depedencies
throught this script,
and cleaned to higher depedencies
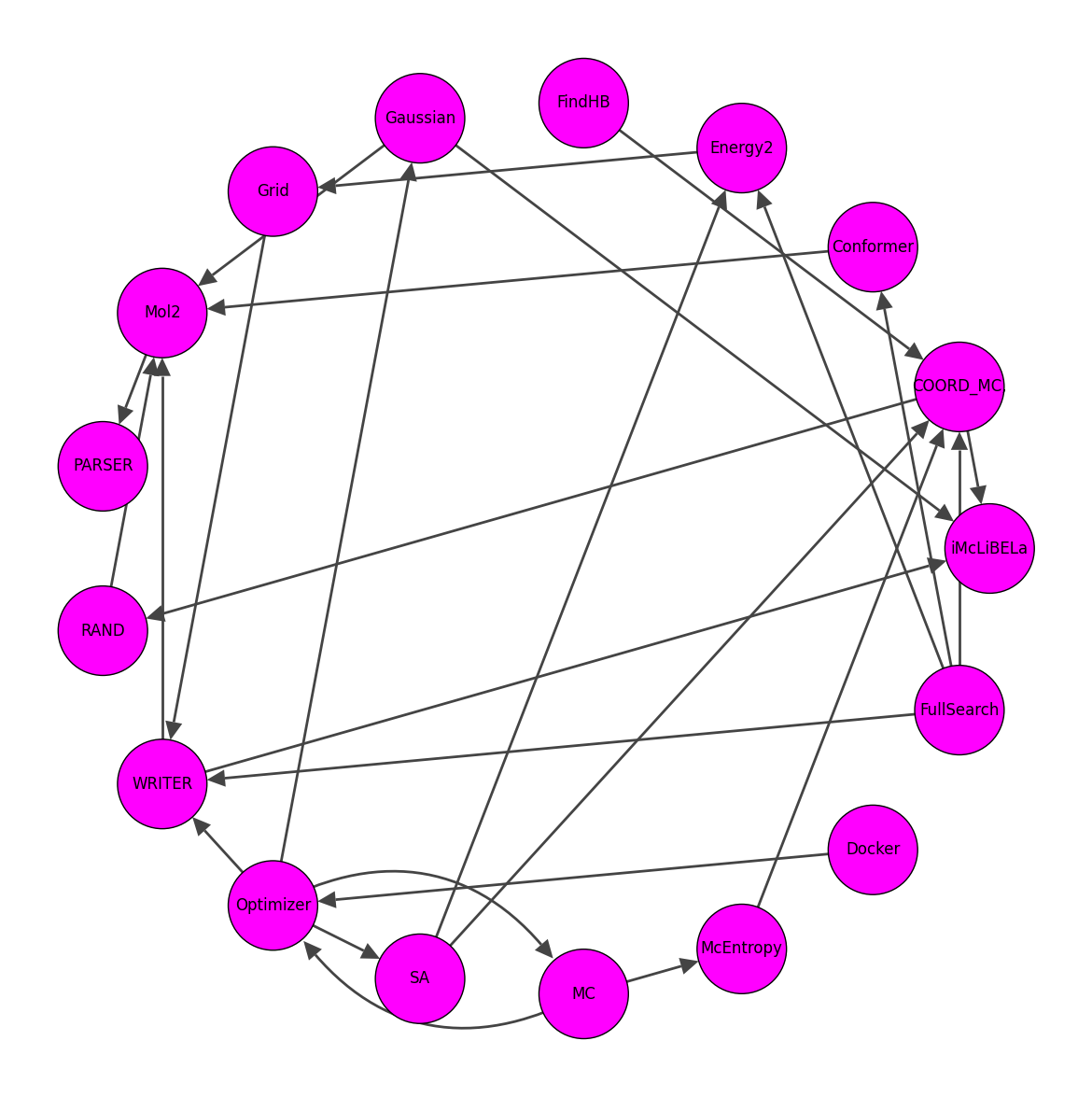
Updated the local and Colab makefiles
- Now we can make one class at a time
Adapting McEntropy, MC and SA classes to LiBELa
- The classes work on the local machine
Notes:
-
In the MC class, I adapted the info variable as in Grid
-
In the MC class, the following error appeared
pyLiBELa/src/pyMC.cpp: In member function ‘void MC::run(Mol2*, Mol2*, Mol2*, std::vector<std::vector<double> >, PARSER*, double)’: pyLiBELa/src/pyMC.cpp:1015:23: warning: format ‘%f’ expects argument of type ‘double’, but argument 3 has type ‘long double’ [-Wformat=] 1015 | sprintf(info, "Boltzmann-weighted average energy: %10.3f kcal/mol @ %7.2f K", this->Boltzmann_weighted_average_energy, T); | ^~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~ ~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~ | | | long doubleand was fixed by changing in the line 1015 the %10.3f to a %10.3Lf, so that is has type long double (Lf).
This also appeared
pyLiBELa/src/pyMC.cpp: In constructor ‘MC::MC(Mol2*, PARSER*, WRITER*)’:
pyLiBELa/src/pyMC.cpp:73:19: warning: format ‘%d’ expects argument of type ‘int’, but argument 3 has type ‘size_t’ {aka ‘long unsigned int’} [-Wformat=]
73 | sprintf(info, "Found %d rotatable bonds in ligand %s.", RotorList.Size(), Lig->molname.c_str());
| ^~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~ ~~~~~~~~~~~~~~~~
| |
| size_t {aka long unsigned int}
and was fixed by changing in the line 73 the %d to a %lu, so that is has type long unsigned int.
Adapting Optimizer class to LiBELa
- The following error appeared
pyLiBELa/src/pyDocker.cpp: In member function ‘void Docker::run(Mol2*, Mol2*, Mol2*, std::vector<double>, PARSER*, unsigned int)’:
pyLiBELa/src/pyDocker.cpp:129:31: warning: format ‘%d’ expects argument of type ‘int’, but argument 14 has type ‘double’ [-Wformat=]
129 | sprintf(info, "%5d %-12.12s %-4.4s %-10.3e %-8.3g %-8.3g %-8.3g %-8.2f %-8.3g %3d %2d %2d %.2f", counter, Lig->molname.c_str(), Lig->resnames[0].c_str(), overlay_fmax,
| ^~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~
130 | 0.0, 0.0, 0.0, 0.0, 0.0, -1, overlay_status, 0.0, si);
| ~~~
| |
| double
pyLiBELa/src/pyDocker.cpp: In member function ‘void Docker::run(Mol2*, Mol2*, Mol2*, std::vector<double>, PARSER*, Grid*, unsigned int)’:
pyLiBELa/src/pyDocker.cpp:266:31: warning: format ‘%d’ expects argument of type ‘int’, but argument 14 has type ‘double’ [-Wformat=]
266 | sprintf(info, "%5d %-12.12s %-4.4s %-10.3e %-8.3g %-8.3g %-8.3g %-8.2f %-8.3g %3d %2d %2d %.2f", counter, Lig->molname.c_str(), Lig->resnames[0].c_str(), overlay_fmax,
| ^~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~
267 | 0.0, 0.0, 0.0, 0.0, 0.0, -1, overlay_status, 0.0, si);
| ~~~
| |
| double
it was fixed by changing formatted string in line 29 from
%5d %-12.12s %-4.4s %-10.3e %-8.3g %-8.3g %-8.3g %-8.2f %-8.3g %3d %2d **%2d** %.2f
to
"%5d %-12.12s %-4.4s %-10.3e %-8.3g %-8.3g %-8.3g %-8.2f %-8.3g %3d %2d **%2f** %.2f" and do the same thing in line 266.
- The following error still remains
pyLiBELa/src/pyOptimizer.cpp: In function ‘void init_module_pyOptimizer()’: pyLiBELa/src/pyOptimizer.cpp:1516:76: error: no matches converting function ‘evaluate_energy’ to type ‘double (class Optimizer::*)(class Mol2*, class std::vector<std::vector<double> >)’ 1516 | double (Optimizer::*ee1)(Mol2*, vector<vector<double> >) = &Optimizer::evaluate_energy; | ^~~~~~~~~~~~~~~ pyLiBELa/src/pyOptimizer.cpp:75:6: note: candidates are: ‘static void Optimizer::evaluate_energy(Mol2*, std::vector<std::vector<double> >, energy_result_t*)’ 75 | void Optimizer::evaluate_energy(Mol2* Lig2, vector<vector<double> > new_xyz, energy_result_t* energy_result){ | ^~~~~~~~~ pyLiBELa/src/pyOptimizer.cpp:48:8: note: ‘static double Optimizer::evaluate_energy(Mol2*, std::vector<std::vector<double> >)’ 48 | double Optimizer::evaluate_energy(Mol2* Lig2, vector<vector<double> > new_xyz){ | ^~~~~~~~~ pyLiBELa/src/pyOptimizer.cpp:1517:94: error: no matches converting function ‘evaluate_energy’ to type ‘void (class Optimizer::*)(class Mol2*, class std::vector<std::vector<double> >, struct energy_result_t*)’ 1517 | izer::*ee2)(Mol2*, vector<vector<double> >, energy_result_t*) = &Optimizer::evaluate_energy; | ^~~~~~~~~~~~~~~ pyLiBELa/src/pyOptimizer.cpp:75:6: note: candidates are: ‘static void Optimizer::evaluate_energy(Mol2*, std::vector<std::vector<double> >, energy_result_t*)’ 75 | void Optimizer::evaluate_energy(Mol2* Lig2, vector<vector<double> > new_xyz, energy_result_t* energy_result){ | ^~~~~~~~~ pyLiBELa/src/pyOptimizer.cpp:48:8: note: ‘static double Optimizer::evaluate_energy(Mol2*, std::vector<std::vector<double> >)’ 48 | double Optimizer::evaluate_energy(Mol2* Lig2, vector<vector<double> > new_xyz){ | ^~~~~~~~~ make: *** [Makefile:115: Optimizer] Error 1
it is probably due to the fact that the functions in this class are defined as static, this approach found on the internet didn’t work.
Useful links:
Week 4
Adapting Optimizer class to LiBELa
-
In order to solve the static overload error shown in Week3, I created another function evaluate_energy2 that corresponds to the version with three arguments. So, in pyOptimizer.cpp pyOptimizer.h scripts, the evaluate_energy method with the energy_t parameter was changed to evaluate_energy2.
-
An attempt to solve any remaining static function errors, every function had an added .staticmethod() as below
.def("name_of_function", & CLASS::name_of_function).staticmethod("name_of_function")
Adapting Docker class to LiBELa
- Adapted tha info variable as Grid
- It works in the local machine
Adapting FullSearch class to LiBELa
- Adapted the info variable as in Grid
- Some format errors were solved in lines 43 and 94, changing from “%d” to “%lu”.
- It still shows the following error
pyLiBELa/src/pyFullSearch.cpp: In member function ‘double FullSearch::do_search()’:
pyLiBELa/src/pyFullSearch.cpp:211:1: warning: no return statement in function returning non-void [-Wreturn-type]
211 | }
| ^
Week 5
Updating Makefile
- It takes 3 minutes to build the .o files. It is done in three steps?
make FILES #downloads the source code make OBJECTS #generates the .o files make LINKAGE #generates the .so libraries -
Checking the depedencies between the libraries
-
Added the Docker class to the github Makefile
-
We still have issues with pySA and pyMc. They depend on the Grid class and can’t be compiled.
- The Optimizer class shows the following error when imported:
undefined symbol: _ZN9Optimizer3RecE
Week 6
Updating Makefile
-
The pyLiBELa is now constructed through the Makefile in three steps. But when called, the Makefile doesn’t make these three targets work one after the other.
- When MC is imported, the following error appears:
ImportError: /content/pyMC.so: undefined symbol: _ZN9Optimizer3RecE - When Optimizer is imported, the following error appears:
ImportError: /content/pyOptimizer.so: undefined symbol: _ZN9Optimizer3RecEThere is a need to reevaluate the way static functions are defined in pyOptimizer.
Useful Links
Week 7
Updating Makefile
-
Google Colab Updated its Python version to Python 3.10.11 so we have to adapt our code to it.
-
Try to replicate LiBELa’s original Makefile
-
Now the objects (.o files) are created and linked to static libraries (.so) in a following step.
Static issues Optimizer
-
We still have some issues about static members of pyOptimizer: Rec, RefLig, Grids and Parser. There are issues with class linkage. The methods are compiled, but Python can’t identify the static members on Colab.
-
I chatted a bit about these issues with ChatGPT to gain some insight into this matter.
-
Tried to adapt some policies suggested by Chat GPT and analyse the errors that appeared. It was inconclusive.
-
We solved the issue by creating a get_Rec function as shown in the Bogo to Bogo link below. We now need to create similar functions with RefLig, Parser and Grids.
Useful Links
Week 8
Boost versions
- When we type in the Colab environment
!ls /usr/lib/x86_64-linux-gnu/libboost_python*it shows that it has libboost_python38.so.1.71.0 installed.
- In our local machine, we have libboost_python310.so.1.74.0 and it is promptly shown in
apt-cache search libboost-python - In Colab, this command shows only
libboost-python1.71.0 - Boost.Python Library
Update Makefile
- Added Docker and FullSearch classes to the Makefile
- The class pyOptimizer is now working!! We had issues with the static members Rec, RefLig, Parser, and Grids. In order to solve it, we had to define the functions getRec, getRefLig, getParser and getGrids outside of the Optimizer class. Then, we did the boost wrapper with a appropriate return policy:
.def("get_Rec", get_Rec, return_value_policy<return_by_value>())
Finishing pyLiBELa
-We now have a Working Colab that downloads the source code and builds pyLiBELa from the Makefile.
SB database
- We are now starting to evaluate pyLiBELa`s performance.
- The SB 2021 Docking Database available here.
- The Database has 1,172 protein-ligand complexes each with a directory containing four files. As we are interested only in the structure of the protein and the ligand, I wrote a script that removes everything else reducing its size from 556,9 MB to 165,8 MB. This clean SB was uploaded to my GoogleDrive.
- The pyLiBELa Collab can now acess the SB database so tests can be run.
- The next challenge is to write a script that calculates the Grids for a number of protein-ligand complexes.
Week 9
OpenMP linkage
- According to StackOverflow, we only need to add the -fopenmp flag to link this library.
- Updated the makefile and it works!
Week 10
Molecular visualization
- We need to prepare a Colab environment in which our docking can be seen. The initial development will be done in this Colab.
- The java based py3Dmol can be used and opens the box.pdb file.
- We seek a viewer with the following characteristics: centered on the ligand, showing neighbour residues, highlighting the ligand and with the box around it.
- In order to use the py3Dmol library better, we could convert the SB dataset files to .pdb format. We can do it by using the openbabel library
obabel -imol2 rec.mol2.gz -opdb -O rec.pdb -
We managed to view the ligand, the box, the protein and an inorganic compound that was only noticed when the .pdb file was opened in Pymol. This inorganic compound can be visualized if you manually type the inorganic elements that can be found in a list. If there is an element from that list, it is shown in the spheres mode.
-
In order to manipulate better the chains and the ligand, we need to join the ligand and receptor files and set the ligand to chain B. We can do it through manipulation of the .pdb files with the sed command in linux. By adding the -i flag, it updates the file.
- The .pdb files were joined as follows:
#original file !tail $name_file_rec #to show the last linesgives the output
CONECT 2612 2611 CONECT 2613 2611 2614 2615 CONECT 2614 2613 CONECT 2615 2609 2609 2613 2616 CONECT 2616 2615 CONECT 2617 2604 2618 2619 CONECT 2618 2617 CONECT 2619 2617 MASTER 0 0 0 0 0 0 0 0 2620 0 2620 0 ENDAs we are going to append another pdb file with a different atom count, we need to remove the CONECT lines. That is because in the pdb format, such lines show the bonds between the atoms, and we don’t want to reset the ATOM id as we are looking for a general solution. When we don’t remove these lines, we get anomalies such as
and
These lines can be removed without damage to viewing. We also need to remove the END line, as it marks the end of the structure.
#removing END and CONECT lines
!sed -i '/END\|CONECT/d' $name_file_rec
!tail $name_file_rec
with the following output
ATOM 2612 HE1 HIE A 166 -4.687 14.216 3.620 1.00 0.00 H
ATOM 2613 NE2 HIE A 166 -2.634 13.840 3.194 1.00 0.00 N
ATOM 2614 HE2 HIE A 166 -2.471 13.015 3.789 1.00 0.00 H
ATOM 2615 CD2 HIE A 166 -1.789 14.362 2.290 1.00 0.00 C
ATOM 2616 HD2 HIE A 166 -0.782 14.028 2.092 1.00 0.00 H
ATOM 2617 C HIE A 166 -2.307 14.471 -1.360 1.00 0.00 C
ATOM 2618 O HIE A 166 -2.772 14.918 -2.417 1.00 0.00 O1-
ATOM 2619 OXT HIE A 166 -2.671 13.389 -0.874 1.00 0.00 O1-
ATOM 2620 MAG MAG A 167 4.767 33.735 19.215 1.00 0.00 Mg
MASTER 0 0 0 0 0 0 0 0 2620 0 2620 0
Now we need to change the ligand chain from A to B.
!head $name_file_lig #shows the first lines
We can see that it resets the ATOM count
COMPND 121P.lig
AUTHOR GENERATED BY OPEN BABEL 3.0.0
ATOM 1 P1 LIG A 1 5.150 32.173 22.030 1.00 0.00 P
ATOM 2 O1 LIG A 1 4.768 32.597 23.390 1.00 0.00 O
ATOM 3 O2 LIG A 1 4.164 32.683 21.069 1.00 0.00 O
ATOM 4 O3 LIG A 1 4.834 30.641 22.025 1.00 0.00 O
ATOM 5 C1 LIG A 1 6.782 32.406 21.624 1.00 0.00 C
ATOM 6 H1 LIG A 1 7.016 33.438 21.679 1.00 0.00 H
ATOM 7 H2 LIG A 1 7.394 31.869 22.301 1.00 0.00 H
ATOM 8 P2 LIG A 1 7.419 31.970 20.137 1.00 0.00 P
We use the sed command again to change the letters
#change A to B in file
!sed -i 's/A /B /g' $name_file_lig
!head $name_file_lig
which gives the output
COMPND 121P.lig
AUTHOR GENERATED BY OPEN BABEL 3.0.0
ATOM 1 P1 LIG B 1 5.150 32.173 22.030 1.00 0.00 P
ATOM 2 O1 LIG B 1 4.768 32.597 23.390 1.00 0.00 O
ATOM 3 O2 LIG B 1 4.164 32.683 21.069 1.00 0.00 O
ATOM 4 O3 LIG B 1 4.834 30.641 22.025 1.00 0.00 O
ATOM 5 C1 LIG B 1 6.782 32.406 21.624 1.00 0.00 C
ATOM 6 H1 LIG B 1 7.016 33.438 21.679 1.00 0.00 H
ATOM 7 H2 LIG B 1 7.394 31.869 22.301 1.00 0.00 H
ATOM 8 P2 LIG B 1 7.419 31.970 20.137 1.00 0.00 P
We also need to remove the CONECT lines from the ligand, but ot the END line, as it is the END of this new file.
!tail $name_file_lig
CONECT 39 37 40 41
CONECT 40 39
CONECT 41 39 42 45 45
CONECT 42 41 43 44
CONECT 43 42
CONECT 44 42
CONECT 45 41 41 46
CONECT 46 32 36 36 45
MASTER 0 0 0 0 0 0 0 0 46 0 46 0
END
we will use the same line as before
#Remove CONECT lines from ligand file
!sed -i '/CONECT/d' $name_file_lig
!tail $name_file_lig
which gives the output
ATOM 39 N3 LIG B 1 17.666 32.006 16.826 1.00 0.00 N
ATOM 40 H12 LIG B 1 18.472 31.504 16.485 1.00 0.00 H
ATOM 41 C10 LIG B 1 17.922 32.786 17.906 1.00 0.00 C
ATOM 42 N4 LIG B 1 19.209 32.864 18.275 1.00 0.00 N
ATOM 43 H13 LIG B 1 19.926 32.459 17.692 1.00 0.00 H
ATOM 44 H14 LIG B 1 19.460 33.328 19.135 1.00 0.00 H
ATOM 45 N5 LIG B 1 16.947 33.303 18.638 1.00 0.00 N
ATOM 46 C11 LIG B 1 15.731 33.075 18.102 1.00 0.00 C
MASTER 0 0 0 0 0 0 0 0 46 0 46 0
END
Finally, we need to concatenate the files
name_file_merge = pdb_code + '.pdb'
!cat $name_file_rec $name_file_lig > $name_file_merge
-Now we can manipulate the chains separately and view the following image.
- With the line
view.addStyle({'chain':'A','within':{'distance':'5', 'sel':{'chain':'B'}}}, {'stick': {'colorscheme':'gray'}})we can show atoms from the main chain which are at a 5 Anstrom distance from the ligand (chain B). And the view is the following
we can see that it sometimes doesn’t show the whole residue, only the atoms in the given distance. Showing the whole residue will be next week’s challenge.
pyLiBELa Docking Grids
- When we calculate the energy through a Grid that is smaller than the ligand, it increases a lot. So we need an estimate of the minimum grid size in which all the ligand could fit. Here we have, for example, the 121P pair with a grid that results in a very high energy. We can see that it gets outside of the calculated grid.
- We have a distribution of the greatest interatomic distance in the following histogram
with this code. That means we need to calculate grids with size 30 in order to fit all the ligands.
- We checked the number of threads in the Virtual Machine in Google Colab. By tiping
!cat /proc/cpuinfowe learn that it only has 2 available threads.
Useful Links
Week 11
Molecular visualization
- In order to show the whole residue within a given distance of the ligand, be just neet to add the flag ‘byres’:True.
- We can alter the window size with the height and widht arguments on the view() function.
- The Magnesium in the 121P pair can be showns in a much better way when we set it such as
aminoacids = ['MET','THR','ASN','LYS','SER','ARG','VAL','ALA','ASP','GLU','GLY','PHE','LEU','SER','TYR','CYS','TRP','LEU','PRO','HIS','GLN','ARG','ILE','HIE'] view.setStyle({ 'chain':'A', 'not':{'resn':aminoacids}}, {'sphere':{}})whatever is in the receptor (chain A) and is not an aminoacid is shown in shperes. We added the standard 23 aminoacids and thte ‘HIE’, which is the way the histidine is written in this file. If there is another residue that differs from its standard three letter name, it will be shown in spheres along the chain. To solve it, we only need to add it to the list.
- Our next challenge will be to show only the lateral chain of the residues within a distance from the ligand. Any other development may be found by studying carefull the Useful links from last week.
pyLiBELa on google Colab
- I based my code in my previous undergraduate research program to organize the data.
- In the Colab, I write the Grid calculation data (time per atom, calculated energies, etc) in a .dat or .csv file.
- In order to calculate the time, I need to check if the tima calculated in the timeit library is in seconds. This was verified through this test. We can see that the time shown on the Colab cell is in seconds, and it corresponds numerically with the one printed.
Calculating Grids
- Now that the Colab environment is all set, we need to analyse how the percentual energy error is affected by grid spacing.
- We saw that most ligands would fit in a grid box with size 30 according to this histogram
- The ideal percentual difference would be smaller than 5%. We need to see the average difference for all the ligand from the SB dataset.
- We may use some filters removing, for instance, positive energies diferença perentual (< q 5% é o ideal) - ver a média para todos filtros - se a energia é positiva, tirar tanto a das coordenadas quanto a dos grids
rodei tudo por 7h - teste sb 25 analisar dados
time energy dá quase 0 121P 1.0 Original energy: -237.910 Time grids: 11.15 Points: 30 30 30 Grid energy: 414.375 Time energy: 0.00
121P 0.5
Original energy: -237.910
Time grids: 100.20
Points: 60 60 60
Grid energy: -61.214
Time energy: 0.00
rodei pra 10
troquei parametros e adaptei código
com uma e duas threads Fiz pra 0.4 e pros outros valores acertando os parâmetros iniciais https://docs.google.com/spreadsheets/d/19CZs9mP9e16ygpjpaHuikuSXUSl9sgjryzwWawZ_bpI/edit?usp=sharing
Cluster testing
rodar no cluster (http://nascimento.ifsc.usp.br/wordpress/?page_id=53) cluster: site alessandro
baixar miniconda no cluster DEFEITO ASANA
testar script pra máquina local criando script
Useful links
Week 12
Setting up to run pyLiBELa on the Cluster.
-
Creating a working local version of the pyLiBELa Colab.
- Having some trouble adapting the Makefile to the cluster’s system specifications.
- Wrote a script that does the same as the pyLiBELa Colab.